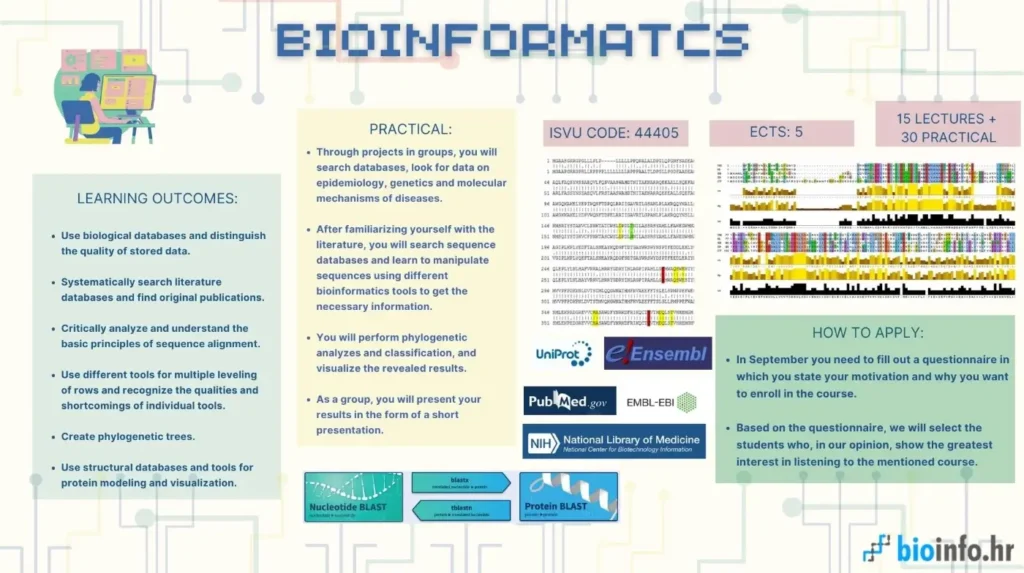
The aim of this course is to equip students with fundamental bioinformatics skills, including the use of biological databases, sequence alignment tools, phylogenetic analysis, and genomic data visualization. Through lectures and hands-on projects, students will learn to analyze epidemiological, genetic, and molecular data, culminating in a comprehensive research experience that integrates sequence analysis, functional genomics, and phylogeny.
The aim of this course is to provide students with a comprehensive understanding of high-throughput sequencing technologies and their applications, emphasizing best practices in experimental design to ensure high-quality data for downstream analysis. Students will gain insights into various sequencing techniques, including whole-genome sequencing, RNA sequencing, methylation analysis, and single-cell sequencing, as well as learn to design experiments for metagenomics, metatranscriptomics, and DNA-protein interaction studies.
This course aims to acquaint biology students with fundamentals of programming and algorithms needed to analyze and solve biological problems by using the R programming language. The course consists of lectures and a practical part in which the students are expected to apply the knowledge gained in classes to solve six groups of exercises. Each student’s solution is graded and used in forming part of the final grade for the class. Each student gets individual feedback with comments for each solved exercise.
The aim of this course is to introduce students to key statistical and machine learning methods for data analysis, with an emphasis on practical applications in the R programming environment. Students will learn techniques such as hypothesis testing, regression, classification, regularization, tree-based methods, support vector machines, clustering, and principal component analysis, gaining skills in both theoretical understanding and hands-on implementation for real-world datasets.
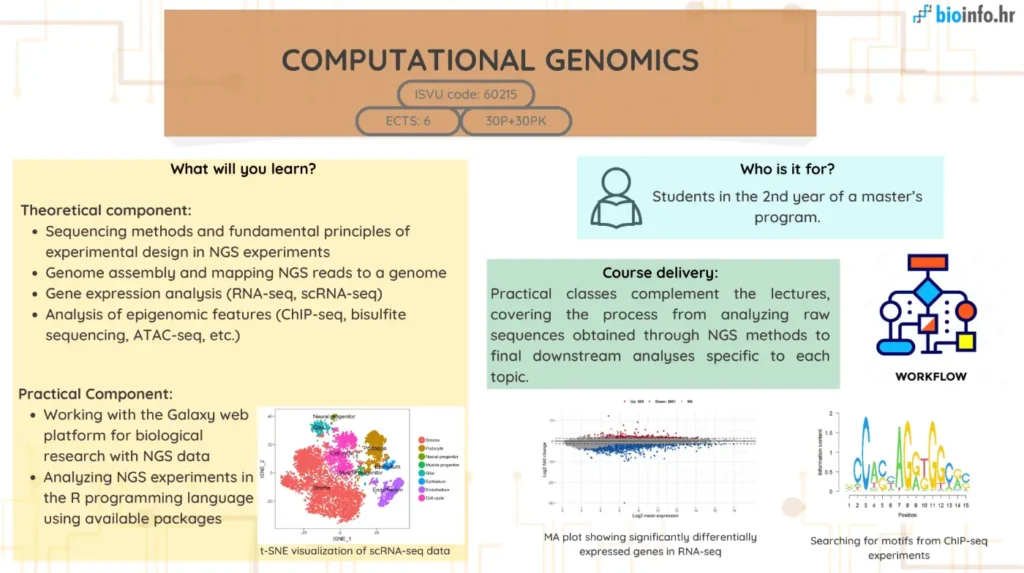
The aim of this course is to provide students with foundational knowledge and practical skills in computational genomics, focusing on next-generation sequencing (NGS) technologies and their applications. Students will explore genome sequencing history, de novo assembly, transcriptome and protein-DNA interaction analysis, and 3D genome structure determination. Through hands-on training, they will learn to use genomic data tools, perform sequence mapping, quality control, and analyze gene expression and ChIP-Seq data using Bioconductor packages in R.
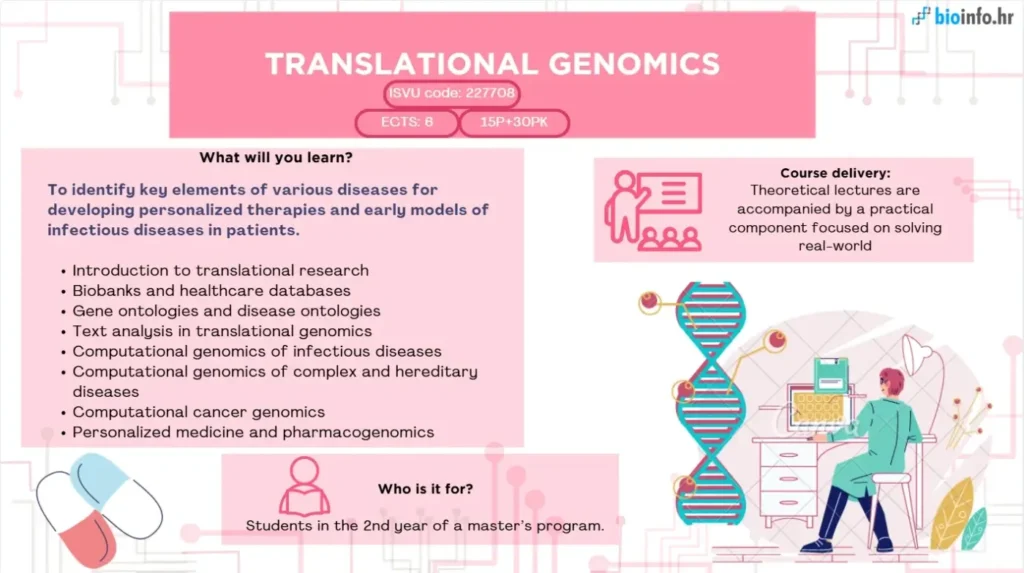
The aim of this course is to provide students with an understanding of translational genomics, focusing on the integration of computational methods with biomedical research to address challenges in diagnosing, characterizing, and treating diseases. Students will explore topics such as health data management, computational genomics of infectious, hereditary, and complex diseases, tumor genomics, gene and disease ontologies, pharmacogenomics, natural language processing in genomics, and the foundations of personalized medicine.

While studying molecular biology at the University of Zagreb he developed a strong interest in applying computational methods and approaches to biosciences. For his master thesis he worked on a problem in X-ray crystallography and developed a program for absorption correction of scattered X-ray data. He obtained his PhD in Bioinformatics/Biochemistry, still pursuing his structural biology interests, working on a computational prediction of structural and physicochemical properties of DNA. He spent 10 years as a research fellow at the International Center for Genetic Engineering and Biotechnology in Trieste, Italy, where he also worked on computational prediction and classification of protein domains using machine learning approaches.
In 2002 he established a computational biology group at the Zagreb University, where he moved permanently in 2006 with the EMBO Young Investigators Programme installation grant, and directed the scientific interests towards the newly emerging field of genomics. His bioinformatics group develops computational tools and uses machine learning techniques to tackle open questions in developmental genomics and metagenomics.
In 2011 he became full professor at the Faculty of Science at Zagreb University. He had several international appointments, including a 4-year adjunct professorship at the University of Oslo, Norway and a two-year adjunct professorship at the University of Skövde, Sweden. From 2008 to 2012 he served as the head of the Division of Biology, Faculty of Science at University of Zagreb and was responsible for managing a division of ~150 staff.
He is involved in teaching four graduate-level courses: Bioinformatics, Algorithms and programming, Statistics and machine learning and Computational genomics. During his time as a group leader more than 30 doctoral and master students graduated under his supervision. Some of his graduates continued with PhD and postdoctoral training at prestigious universities around the world, including LMB Cambridge, UK; EMBL, Heidelberg, Germany; RIKEN, Japan and ETH, Zurich, Switzerland.
His scientific track-record includes more than 60 publications in high-level journals and talks in many renowned institutions throughout the world. His research topics are: developmental and differentiation genomics, metagenomics, population genomics and glycomics, origins of multicellularity, epigenomics of cancer, development of computational methods and application of machine learning in genomics and molecular biology.
He is the reviewer in a number of scientific journals, as well as national and international funding bodies and programmes, such as ESF Programme, EC FP7, H2020, and Horizon Europe Programmes, Estonian National Funding Agency, Hungarian National Funding Agency, Flanders Research Foundation, The Netherlands ZonMw Vidi Programme, Croatian National Funding Agencies. He reviewed program applications for Croatian pre-accession and accession structural funds.
Prof. Vlahoviček is a Fellow of the Academia Europaea, and a member of several professional societies, at the national and international level. Professor Vlahoviček is a strong proponent of science reforms in Croatia and evidence-based policy making. He served in the steering committee of Croatia’s highly successful science funding body, the Unity through Knowledge Fund (UKF) and has participated in several strategy-drafting panels at the university and national level. He also served as the Member of the Steering Committee of the Ruđer Bošković Institute, a leading Croatian research entity in natural sciences, with ~900 staff.